Data Packaging
Every Brain-Score benchmark requires experimental data packaged in standardized formats. This tutorial covers how to prepare your stimuli and measurements for use in benchmarks.
Overview
Every data plugin is built around two fundamental data structures:
- Stimulus Sets: Collections of experimental stimuli with metadata
- Data Assemblies: Multi-dimensional arrays containing experimental measurements
Raw Data → Organize Stimuli → Create Assembly → Validate → Package → Register
The Data Structures
StimulusSet
StimulusSet extends the pandas DataFrame class with functionality for handling stimulus files:
# Located: core/brainscore_core/supported_data_standards/brainio/stimuli.py
from brainscore_core.supported_data_standards.brainio.stimuli import StimulusSet
# Create stimulus metadata
stimuli_data = [
{'stimulus_id': 'img_001', 'category': 'face', 'emotion': 'happy'},
{'stimulus_id': 'img_002', 'category': 'face', 'emotion': 'sad'},
{'stimulus_id': 'img_003', 'category': 'object', 'subcategory': 'car'},
{'stimulus_id': 'img_004', 'category': 'object', 'subcategory': 'house'},
...
]
# Create StimulusSet
stimulus_set = StimulusSet(stimuli_data)
# Link to actual files
stimulus_set.stimulus_paths = {
'img_001': '/path/to/face_happy.jpg',
'img_002': '/path/to/face_sad.jpg',
'img_003': '/path/to/car.jpg',
'img_004': '/path/to/house.jpg',
...
}
# Set identifier
stimulus_set.name = 'MyExperiment2024'
If you already have a CSV file that contains all relevant stimulus information, you can convert a Pandas DataFrame into a StimulusSet
# Located: core/brainscore_core/supported_data_standards/brainio/stimuli.py
import pandas as pd
from brainscore_core.supported_data_standards.brainio.stimuli import StimulusSet
# Read CSV
df = pd.read_csv("stimuli.csv")
# Note: stimulus_id must remain as a column (not index)
# Convert stimulus_id to string for compatibility with packaging code
df['stimulus_id'] = df['stimulus_id'].astype(str)
# Rename 'filename' to 'image_filename' to avoid conflict with packaging code
# The packaging code will add its own 'filename' column with zip filenames
if 'filename' in df.columns:
df = df.rename(columns={'filename': 'image_filename'})
# Create StimulusSet
stimulus_set = StimulusSet(df)
# Set identifier
stimulus_set.name = 'MyExperiment2024'
# Create stimulus_paths dictionary mapping stimulus_id to file paths
# This is required for packaging
# The keys must match the stimulus_id values in the DataFrame (as strings)
# Use 'image_filename' if available, otherwise fall back to 'filename'
filename_col = 'image_filename' if 'image_filename' in stimulus_set.columns else'filename'
stimuli_directory = "/path/to/stimuli"
stimuli_dir = Path(stimuli_directory)
stimulus_set.stimulus_paths = {
str(row['stimulus_id']): stimuli_dir / row[filename_col]
for _, row in stimulus_set.iterrows()
}
Essential Components:
| Component | Description | Required |
|---|---|---|
stimulus_id |
Unique STRING identifier for each stimulus | Yes |
stimulus_paths |
Mapping from IDs to file locations | Yes (for file-based stimuli) |
name |
Unique name for the stimulus set | Yes |
| Metadata columns | Experimental variables (category, condition, etc.) | Recommended |
⚠️ Note: A 'filename' field cannot be part of the StimulusSet. To avoid conflict with packaging code, rename 'filename' to 'stimulus_filename'. The packaging code will add its own 'filename' column with zip filenames.
Naming Conventions
Use consistent naming for your data identifiers:
| Component | Convention | Example |
|---|---|---|
| Base identifier | AuthorYear or descriptive |
MajajHong2015, ImageNet |
| Sub-datasets | Separate with dot (.) |
MajajHong2015.IT, MajajHong2015.V4 |
| Multiple words | Use underscore (_) |
Malania2007.vernier_only |
# Examples
stimulus_set.name = 'MyExperiment2024' # Base identifier
stimulus_set.name = 'MyExperiment2024.condition_A' # Sub-dataset variant
Tip: The same identifier is typically used for both the StimulusSet and its corresponding DataAssembly, keeping them linked.
Packaging StimulusSet Locally
Packaging the StimulusSet will produce two files: The first is a CSV file containing the stimulus metadata (the DataFrame) - stimulus IDs, categories, conditions, etc. The second is a ZIP file containing the actual stimulus files (images, audio, etc.). The enclosed stimuli are renamed to based on the stimulus_id and referenced by stimulus_paths.
# Located: core/brainscore_core/supported_data_standards/brainio/packaging.py
from brainscore_core.supported_data_standards.brainio.packaging import package_stimulus_set_locally
package_stimulus_set_locally(
proto_stimulus_set=stimulus_set,
stimulus_set_identifier=stimulus_set.name,
)
Output:
{
'identifier': 'MyExperiment2024',
'csv_path': '/path/to/Downloads/brainscore_packages/stimulus_MyExperiment2024.csv',
'zip_path': '/path/to/Downloads/brainscore_packages/stimulus_MyExperiment2024.zip',
'csv_sha1': '1d47ea4a09ddd72cebabca95b985646650f21646',
'zip_sha1': 'c96036d459f0a2ce4494ba73a2b18b8eec59f6b6'
}
When performing local packaging, unless a path is specified, the StimulusSet will be stored in ~/Downloads/brainscore_packages/. The csv_sha1 and zip_sha1 are the hashes for the respective files. The hashes are used to ensure data integrity and identify the exact version of each file.
⚠️ Note: You will need the hashes when registering the data plugin, so keep them safe.
DataAssembly
DataAssembly is built on xarray for multi-dimensional scientific data:
# Located: core/brainscore_core/supported_data_standards/brainio/assemblies.py
from brainscore_core.supported_data_standards.brainio.assemblies import DataAssembly
import numpy as np
# Create data array: 100 stimuli × 50 recording sites
data = np.random.rand(100, 50)
assembly = DataAssembly(
data,
coords={
'stimulus_id': ('presentation', [f'img_{i:03d}' for i in range(100)]),
'measurement_id': ('measurement', [f'unit_{i}' for i in range(50)]),
'condition': ('presentation', ['A'] * 50 + ['B'] * 50)
},
dims=['presentation', 'measurement']
)
The Coordinate System:
Coordinates provide named labels for dimensions. The format is:
'coordinate_name': ('dimension_name', [values])
| Coordinate Type | Purpose | Example |
|---|---|---|
| Identification | Unique identifiers | stimulus_id, neuroid_id |
| Grouping | Experimental conditions | condition, category |
| Linking | Connect stimuli to responses | stimulus_id (shared across assemblies) |
| Metadata | Experimental parameters | subject, session, trial |
Specialized Assembly Types
NeuroidAssembly (Neural Recordings)
For neural data with dimensions: presentations × neuroids × time_bins
# Located: core/brainscore_core/supported_data_standards/brainio/assemblies.py
from brainscore_core.supported_data_standards.brainio.assemblies import NeuroidAssembly
# Neural data: presentations × neuroids × time_bins
# This is just a random example
neural_data = np.random.rand(200, 100, 10)
assembly = NeuroidAssembly(
neural_data,
coords={
# 'coordinate_name': ('dimension_name', [values])
# Presentation coordinates
# e.g., `stimulus_id` is a coordinate attached the the `presentation` dimension
'stimulus_id': ('presentation', [f'img_{i:03d}' for i in range(200)]),
'repetition': ('presentation', [i % 3 for i in range(200)]),
# Neuroid coordinates
'neuroid_id': ('neuroid', [f'neuron_{i:03d}' for i in range(100)]),
'region': ('neuroid', ['V4'] * 50 + ['IT'] * 50),
'animal': ('neuroid', ['monkey_A'] * 100),
# Time coordinates
'time_bin_start': ('time_bin', list(range(0, 100, 10))),
'time_bin_end': ('time_bin', list(range(10, 110, 10))),
},
dims=['presentation', 'neuroid', 'time_bin']
)
# Add experimental metadata
assembly.attrs['experiment_date'] = '2024-03-22'
assembly.attrs['sampling_rate'] = 280000 # Hz
Standard Coordinates for Neural Data:
| Dimension | Description | Required Coordinates | Optional Coordinates |
|---|---|---|---|
| presentation | A single instance of showing a stimulus | stimulus_id |
repetition, session, condition |
| neuroid | A single unit of neural measurement | neuroid_id |
region, animal, electrode, hemisphere |
| time_bin | A temporal window of the neural measurement | time_bin_start, time_bin_end |
— |
Optional Coordinate Use Cases:
| Coordinate | Use Case |
|---|---|
region |
Allows packaging multiple brain regions in one assembly, then slicing (e.g., assembly.sel(region='IT')) — though separate assemblies per region is often cleaner |
repetition |
Enables split-half ceiling calculations by grouping repeated presentations of the same stimulus |
animal |
Enables cross-subject analyses or leave-one-subject-out ceiling calculations |
electrode |
Useful for tracking which physical electrode recorded each neuroid |
hemisphere |
Allows hemisphere-specific analyses (e.g., assembly.sel(hemisphere='left')) |
session |
Allows filtering or grouping data by recording session for session-level analyses |
condition |
Enables slicing data by experimental condition (e.g., assembly.sel(condition='attended')) |
| among others.... |
BehavioralAssembly (Behavioral Responses)
For behavioral measurements, typically with dimension: presentations
# Located: core/brainscore_core/supported_data_standards/brainio/assemblies.py
from brainscore_core.supported_data_standards.brainio.assemblies import BehavioralAssembly
# Simple 1D behavioral data
behavioral_data = np.array([1, 0, 1, 1, 0, 1, 0, 0, 1, 1]) # correct/incorrect
assembly = BehavioralAssembly(
behavioral_data,
coords={
'stimulus_id': ('presentation', [f'img_{i:03d}' for i in range(10)]),
'subject': ('presentation', ['subj_01'] * 10),
'condition': ('presentation', ['easy'] * 5 + ['hard'] * 5),
'response_time': ('presentation', [0.5, 0.7, 0.4, 0.8, 1.2, 0.9, 1.5, 1.1, 0.6, 0.8]),
'correct': ('presentation', [1, 0, 1, 1, 0, 1, 0, 0, 1, 1])
},
dims=['presentation']
)
For multi-choice tasks (probability distributions):
# Probability distribution over choices
choice_data = np.array([
[0.8, 0.1, 0.1], # img_001; 80% choice A, 10% B, 10% C
[0.2, 0.7, 0.1], # img_002;
[0.1, 0.2, 0.7] # img_003
])
assembly = BehavioralAssembly(
choice_data,
coords={
'stimulus_id': ('presentation', ['img_001', 'img_002', 'img_003']),
'choice': ('choice', ['A', 'B', 'C'])
},
dims=['presentation', 'choice']
)
Standard Coordinates for Behavioral Data:
| Dimension | When Used | Typical Coordinates |
|---|---|---|
presentation |
Always (links responses to stimuli) | stimulus_id (required), subject, condition, trial |
choice |
For probability distributions over options | choice (the option labels) |
Key Difference from NeuroidAssembly:
BehavioralAssemblyhas no enforced dimensions. UnlikeNeuroidAssemblywhich validates that dimensions are exactlypresentation × neuroid(±time_bin),BehavioralAssemblyis intentionally flexible because behavioral experiments vary widely in structure.
Behavioral data comes in many forms:
| Data Type | Dimensions | Example |
|---|---|---|
| Binary responses | presentation |
Correct/incorrect (1D array) |
| Probability distributions | presentation × choice |
Softmax over N classes |
| Reaction times | presentation |
Response latencies |
| Similarity judgments | presentation |
Odd-one-out choices |
As mentioned in the What is a Benchmark tutorial, this flexibility extends to benchmarks: there is no BehavioralBenchmark helper class (unlike NeuralBenchmark). Each behavioral benchmark inherits directly from BenchmarkBase and implements its own __call__ method to handle its specific task logic.
Optional Coordinate Use Cases:
| Coordinate | Use Case |
|---|---|
subject |
Enables cross-subject ceiling calculations and leave-one-subject-out analyses |
condition |
Allows slicing by experimental condition (e.g., assembly.sel(condition='hard')) |
trial |
Tracks trial order for sequence-dependent analyses |
repetition |
Groups repeated presentations for reliability calculations |
response_time |
Stores reaction times for RT-based analyses |
correct |
Stores ground truth for accuracy calculations |
Packaging DataAssembly Locally
# Located: core/brainscore_core/supported_data_standards/brainio/packaging.py
from brainscore_core.supported_data_standards.brainio import packaging
packaging.package_data_assembly_locally(
proto_data_assembly=assembly,
assembly_identifier=stimulus_set.name, # We use the same stimulusSet name for the assembly
stimulus_set_identifier=stimulus_set.name,
assembly_class_name="NeuroidAssembly", # For most neural data, use NeuroidAssembly. For behavioral data, use BehavioralAssembly
)
Other supported assembly class types can be found in core/brainscore_core/support_data_standards/brainio/fetch.py:resolve_assembly_class(). These include DataAssembly, NeuroidAssembly, BehavioralAssembly, PropertyAssembly, MetadataAssembly, and SpikeTimesAssembly.
Output:
{
'identifier': 'MyExperiment2024',
'path': '/path/to/Downloads/brainscore_packages/assy_stimulus_MyExperiment2024.nc',
'sha1': '3c45d95e2e61616c885758df0d463fddbfc2b427',
'cls': 'NeuroidAssembly'
}
When performing local packaging, unless a path is specified, the dataAssembly will be stored in ~/Downloads/brainscore_packages/. The sha1 is the hash for the assembly. The hashes are used to ensure data integrity and identify the exact version of each file.
⚠️ Note: Like the StimulusSet, you will need the hash when registering the data plugin, so keep it safe.
The Data Packaging Pipeline
Step 1: Prepare Stimulus Set
Create a StimulusSet with metadata and link it to your stimulus files. Ensure each stimulus has a unique stimulus_id and set the name attribute.
stimulus_set = StimulusSet(metadata_df)
stimulus_set.name = 'MyExperiment2024'
stimulus_set.stimulus_paths = {stim_id: path for stim_id, path in ...}
Step 2: Create Data Assembly
Build a DataAssembly with your experimental data. Include stimulus_id as a coordinate to link responses to stimuli.
assembly = DataAssembly(data, coords={...}, dims=['presentation', 'neuroid', 'time_bin']) # for NeuroidAssembly
Step 3: Validate and Package
Verify your data has unique IDs, no missing files, and no unexpected NaN values. Then package locally using package_stimulus_set_locally() and package_data_assembly_locally() — save the output hashes. The packaging function will perform some validation.
Step 4: Upload
Once your data is packaged locally, upload it to Brain-Score's S3 storage through the Brain-Score website. Navigate to the upload page through the Central Profile Page and submit your packaged files (CSV, ZIP, and NC files).
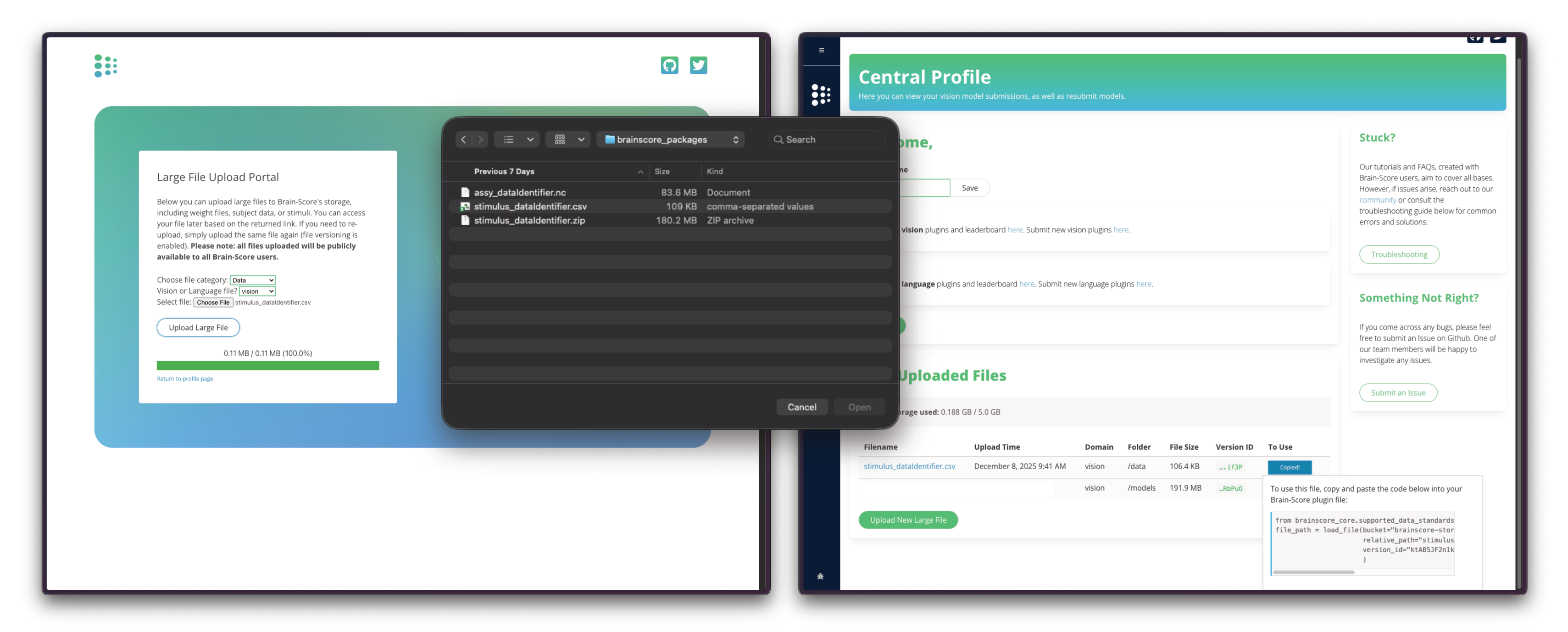
After uploading your three files, you'll receive the S3 bucket path and version_id values needed for registration through the Copy Code button.
The Upload Portal will be soon updated to provide the exact code for loading the stimulus set and data assemblies.
Step 5: Register in __init__.py
The __init__.py file registers your data with Brain-Score's plugin system. When users call load_stimulus_set('MyExperiment2024') or load_dataset('MyExperiment2024'), these registrations tell the system where to find and how to load your data from S3.
# vision/brainscore_vision/data/myexperiment2024/__init__.py
from brainscore_vision import data_registry, stimulus_set_registry, load_stimulus_set
from brainscore_core.supported_data_standards.brainio.s3 import load_stimulus_set_from_s3, load_assembly_from_s3
from brainscore_core.supported_data_standards.brainio.assemblies import NeuroidAssembly
# Provide bibtex for proper citation of the original dataset
BIBTEX = """@article{MyExperiment2024,
title = {Neural correlates of visual object recognition in primates},
volume = {42},
doi = {10.1234/neuroscience.2024.00123},
journal = {Journal of Neuroscience},
author = {Pradeepan, Kartik S., and Ferguson, Mike.},
year = {2024},
}"""
# Register stimulus set
# The lambda ensures lazy loading — data is only fetched when actually requested
stimulus_set_registry['MyExperiment2024'] = lambda: load_stimulus_set_from_s3(
identifier="MyExperiment2024", # Must match the key in the registry
bucket="brainscore-storage/brainscore-vision/data/..." # S3 bucket path (provided after upload)
csv_sha1="54d0645a875fcfb29bdf414db9cb54cc0ab3cacf", # From local packaging output
zip_sha1="c96036d459f0a2ce4494ba73a2b18b8eec59f6b6", # From local packaging output
csv_version_id="U405jdh2ECWCzwoZauxh0VNDSAHAb.2s", # S3 version ID (provided after upload)
zip_version_id="cTVQxPGpom_seCpwN2ltG_LK7eYGbZid", # S3 version ID (provided after upload)
filename_prefix="stimulus_" # Prefix used during packaging
)
# Register data assembly
data_registry['MyExperiment2024'] = lambda: load_assembly_from_s3(
identifier="MyExperiment2024",
version_id="yusgi5xpyrNzU10cjk69Z49G.CSyujXO", # S3 version ID
sha1="ef217247308f806c2435f452c53481a04f5a6ba3", # From local packaging output
bucket="brainscore-storage/brainscore-vision/...", # S3 bucket path (provided after upload)
cls=NeuroidAssembly, # Assembly class (NeuroidAssembly or BehavioralAssembly)
stimulus_set_loader=lambda: load_stimulus_set('MyExperiment2024') # Links to the stimulus set
)
Note: The
bucketandversion_idvalues are provided by the Brain-Score team after you upload your packaged data. Thesha1hashes come from your local packaging output.
Step 6: Create Tests
Every data plugin should include a test.py file to validate data loading, structure, and alignment. Tests marked with @pytest.mark.private_access are skipped in CI pull request builds (which can't access private S3 data) but run locally and in post-merge CI.
# vision/brainscore_vision/data/myexperiment2024/test.py
import numpy as np
import pytest
from brainscore_vision import load_dataset, load_stimulus_set
@pytest.mark.private_access
class TestStimulusSet:
def test_stimulus_set_exists(self):
"""Test that stimulus set loads correctly"""
stimulus_set = load_stimulus_set('MyExperiment2024')
assert stimulus_set is not None
assert stimulus_set.identifier == 'MyExperiment2024'
def test_stimulus_set_counts(self):
"""Test expected number of stimuli"""
stimulus_set = load_stimulus_set('MyExperiment2024')
assert len(stimulus_set) == 100 # Replace with your expected count
assert len(np.unique(stimulus_set['stimulus_id'].values)) == 100
@pytest.mark.private_access
class TestAssembly:
def test_assembly_exists(self):
"""Test that assembly loads correctly"""
assembly = load_dataset('MyExperiment2024')
assert assembly is not None
assert assembly.identifier == 'MyExperiment2024'
def test_assembly_structure(self):
"""Test assembly has required dimensions and coordinates"""
assembly = load_dataset('MyExperiment2024')
# Check dimensions
assert 'presentation' in assembly.dims
# Check required coordinates (handles MultiIndex)
assert 'stimulus_id' in assembly.coords or 'stimulus_id' in assembly.indexes
def test_assembly_alignment(self):
"""Test stimulus set and assembly are properly linked"""
assembly = load_dataset('MyExperiment2024')
stimulus_set = assembly.stimulus_set
# All assembly stimulus IDs should exist in stimulus set
assembly_stimuli = set(assembly['stimulus_id'].values)
stimulus_set_stimuli = set(stimulus_set['stimulus_id'].values)
assert assembly_stimuli.issubset(stimulus_set_stimuli)
Running Tests:
# Run all tests for your data plugin
pytest vision/brainscore_vision/data/myexperiment2024/test.py -v
⚠️ Note: There are various ways to structure your tests. You can see more examples in every data plugin in
vision/brainscore_vision/data/{data identifier}/test.py.
Data Plugin Directory Structure
vision/brainscore_vision/data/myexperiment2024/
├── __init__.py # Registration
├── test.py # Unit tests
├── requirements.txt # Dependencies (optional)
└── data_packaging/ # Packaging scripts
└── data_packaging.py # How data was packaged
⚠️ Note: Once you've built the
__init__.pyandtest.py, you're almost done. Place all files in an appropriately named folder. While not manadatory for the function of your benchmark, including the code used to process and package your data is highly recommended. This will help reproduce your benchmark as well as address any scientific questions.
Data Quality Checklist
Before packaging data, verify:
| Check | How to Verify |
|---|---|
| Unique stimulus IDs | assert stimulus_set['stimulus_id'].nunique() == len(stimulus_set) |
| All files exist | Loop through stimulus_paths and check Path(path).exists() |
| No NaN values | assert not np.isnan(assembly.values).any() |
| Consistent coordinates | Verify stimulus_id values match between StimulusSet and DataAssembly |
| Proper dimensions | Check that assembly dims match expected structure |
Common Issues and Solutions
Problem: "Duplicate stimulus IDs"
# Solution: Ensure unique IDs
assert stimulus_set['stimulus_id'].nunique() == len(stimulus_set)
Problem: "Stimulus paths not found"
# Solution: Verify all paths exist
for stimulus_id, path in stimulus_set.stimulus_paths.items():
if not Path(path).exists():
print(f"Missing: {stimulus_id} -> {path}")
Problem: "Assembly dimension mismatch"
# Solution: Check dimensions
print(f"Assembly dims: {assembly.dims}")
print(f"Expected: ['presentation', 'neuroid'] or ['presentation']")
Problem: "stimulus_id type mismatch"
The stimulus_id must be a STRING type. Integer IDs will cause issues with packaging and alignment.
# Solution: Convert stimulus_id to string
df['stimulus_id'] = df['stimulus_id'].astype(str)
stimulus_set = StimulusSet(df)
# Also ensure assembly stimulus_ids are strings
coords = {'stimulus_id': ('presentation', [str(id) for id in stimulus_ids]), ...}
Problem: "'filename' column conflict"
The packaging code adds its own filename column. If your data already has one, it will conflict.
# Solution: Rename 'filename' before creating StimulusSet
if 'filename' in df.columns:
df = df.rename(columns={'filename': 'image_filename'})
Problem: "Stimulus IDs don't match between StimulusSet and Assembly"
All stimulus_id values in the assembly must exist in the StimulusSet.
# Solution: Check alignment
stim_ids = set(stimulus_set['stimulus_id'])
assembly_ids = set(assembly.coords['stimulus_id'].values)
missing = assembly_ids - stim_ids
if missing:
print(f"Assembly has IDs not in StimulusSet: {missing}")
Problem: "NaN values in assembly"
Unexpected NaN values can cause metric calculations to fail.
# Solution: Check for NaN values
nan_count = np.isnan(assembly.values).sum()
if nan_count > 0:
print(f"Warning: {nan_count} NaN values found")
# Either fix the source data or document that NaNs are expected
Next Steps
Once your data is packaged:
- Neural Benchmarks — Create benchmarks for neural recording data
- Behavioral Benchmarks — Create benchmarks for behavioral data


